Grain Size Project
CRISPR-based genome editing of grain size regulators for novel variation to increase wheat genetic yield potential
- PD: Wanlong Li Institution: South Dakota State University
- Co-PD: Bing Yang Institution: Iowa State University
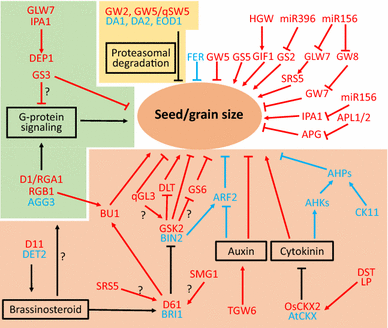
Grain size (GS) is the major driver for further growth of wheat yield. Much progress has been made in dissecting the genetic network regulating GS in the model plants rice and Arabidopsis, wherein more than half of the underlying genes are negative GS regulators or negatively regulated by microRNAs. But little is known about GS regulation in wheat. This knowledge gap significantly limits our effort to improve wheat yield. Hypothesizing that conserved genetic pathways underlie GS variation in rice and wheat, we teamed up with expertise in wheat genetics/genomics and genome editing to address Area Priority 1 of this NIFA-IWYP program (A1142) by creating novel variation of the negative GS regulators using CRISPR-based technology for significant increase of genetic yield potential. Our objectives include:
- Develop an improved CRISPR/Cas9 system for editing GS candidate genes;
- Identify mutations in the GS candidate genes;
- Characterize the effect of mutations on GS; and
- Transfer beneficial mutations into elite durum wheat.
We have optimized the CRISPR/Cas9 system for high-efficiency genome editing in wheat, identified 32 GS candidate genes in wheat genome, and set up platforms for high-throughput GS phenotyping and for early-stage mutation detection. We expect to develop an improved CRIPSR/Cas9 system for wheat with enhanced editing efficiency, targeting flexibility and accuracy, a panel of novel GS mutations and associated knowledge, a package of novel germplasm with enhanced grain yield potential and functional markers. Thus, this project will significantly contribute to IWYP’s goal to increase wheat yield by 50% by 2034.
Primers used in this study
| Gene name | Primer name (F/R) | Forward primer (5`-> 3`) | Reverse primer (5`-> 3`) |
|---|---|---|---|
| APG-1AS | WL3971/WL3972 | GAGTTGGTGCGTGCGCTA | GGACTCGACGGACTTGCTT |
| APG-1BS | WL3973/WL3974 | CCGTGCTCTTCCTTTTCTTC | ACCATGTGTCCAGCGCTTT |
| APG-1DS | WL4092/WL4093 | TGGTGCGCTACTCCTCCT | GGTTTCTTGCGAGGGTTGAC |
| BAS1-3A | WL4234/WL4276 | CACAGCTCAACGAGTGAGTGAGT | CTGGGGACGTATGTATGGATGT |
| BAS1-3B | WL4236/WL4237 | CTATCACAGGACGCCAGG | CGAAGAAGAGACAACTCCAAGGT |
| BAS1-3DS | WL4238/WL4239 | CTCTGTAGCACGCCGCAA | TGGGAGATCAAGCAAACAAA |
| CKX2-3.1-3AS | WL3901/WL3902 | CAACGCACGGACACTTAGC | CGTGTGCGTACGTAGAAGATGAG |
| CKX2-3.1-3BS | WL4101/WL3904 | TCCTGTGCTGTGTACCAGAG | ATGAACGTGATTCCACGCC |
| CKX2-3.1-3DS | WL3905/WL3906 | CATTTCATTTATCATGACTTCCC | GTGCGTGCTTGCCCATATC |
| CKX2-3.2-3BS | WL4082/WL3908 | CTTTGTGTGTGTAATTTCTCCCG | GCTCTAATTAAATACGAAGCGTG |
| CKX2-3.2-3DS | WL3909/WL3910 | AAGAATCTTTCGTCGGTCCT | GGTGAACGAGTTGTGGTTACTTA |
| CKX2-3.3-3AS | WL3911/WL3912 | TACCTACGCAAGCAGGATGGA | GACGAAGACGACGATGACG |
| CKX2-3.3-3BS | WL3913/WL3914 | TTGCAAGCCCCAATAACG | ACAGCGTCGAACAGGTCA |
| CKX2-3.3-3DS | WL3915/WL3916 | GGGTCCCATGTAGTTTACCTTTA | TAGGCACCATAGGCACGG |
| CKX2-3.5-3AS | WL3919/WL3920 | AAGACTCACAGGAAGCAAGCTAA | CGCAAACAAAAGATGATACTCAC |
| CKX2-3.5-3BS | WL3921/ WL3922 | ACCATCCGAAAGAAAACGAA | GCATTAGTACGTGAAGTCGTGAA |
| CKX2-3.5-3DS | WL4056/WL4057 | CGAACCAGATGGATCCGAG | CCATTAGTACGTGAACGTTGGC |
| DEP1-5AL | WL4063/WL4064 | CGTCCGCCCTTGATTTTC | GACCACTAGCCTAGATGCAGGAG |
| DEP1-5BL | WL4003/WL4004 | TGCACTCGCACTCTCTCTCTAAA | CAGCACCAACAACTGCTACTACT |
| DEP1-5DL | WL4143/WL4144 | GCATTTGTTTCCTTGGCAC | GCTGCCGAGATAAATAACCCTA |
| EOD1-4AS | WL4161/WL4162 | TGACAAAAGGACCACTTCTGCT | CAAACACACTAAAGCACGTCA |
| EOD1-4BL | WL4163/WL4164 | GTTCCATAGCCGAGGCCC | TGATGTGCTGGTGAGCTTTC |
| EOD1-4DL | WL4165/WL4166 | TTAGAAGAGAGAACCTTTTTAGGAC | AAAGCCACAAGCCACGTAAT |
| GL3-5AS | WL4062/WL3948 | ATCGTTTGTGGTTGACCTTTC | AGCTAGAAAGTACTGCCGCC |
| GL3-5DS | WL4066/WL3952 | AGAACTGCCCATGTGACAA | AAGGTGAGCAGACTTAGACCGT |
| GLW7-2AS | WL3935/WL3936 | TATCAATGTTGTGCCTGTTTCT | CCAAGTGAAAACCAAATGCTC |
| GLW7-2BS | WL4047/WL4048 | GCAAGATAAAAACTGGAGG | GGCGATAAGGTTTATAAGG |
| GS2-6AL | WL4069/WL3960 | GATTAATCCTGCAAATCTAGCTG | AAAGACAAGGGCTACTGTGC |
| GS2-6BL | WL3961/WL3962 | TAGCAGCGTCCCTCCTCCGAA | AGGGCTACTGTATGGCATGG |
| GS2-6DL | WL3963/WL3964 | CCTCCTTGTTTGTTTTGCCG | CATCTTGGCTTGGCTGTCTT |
| GW2-6AS | WL4007/WL4008 | CGTGTCACAAAACTAATTGGG | CTACGGCAGAACAAATGCAA |
| GW2-6BS | WL4009/WL4010 | GGGCCAGCAGCAGAGAGA | CAAACACAGGCACCTAGCAC |
| GW2-6DS | WL4011/WL4012 | GACATCATACAAGTGGGGAAGGA | TACGGCAGAACAAATGCAAC |
| GW7-2DS | WL3977/WL4046 | GCATGTTCTATCTATGAGCGAGT | CGACCTAAACTTTGGGAATCTAC |
| GW7-2AS | WL3979/WL3980 | CTAGCTTCTTGCCCGCAG | GTGGTTGTTTCTTACTGGACCTT |
| GW7-2BS | WL3981/WL3982 | CCGTCCTCTCCTCTCCTCT | ACAAGGGAAAAACCACTTGATA |
| GW8-5AL | WL3953/WL3954 | CTCGCATCATAAGAATGGAAG | TGGCAGTTCATCTCGTTGTC |
| GW8-5BL | WL3955/WL3956 | AAATTCAGGGACAAGGTTCG | TGCGGTTATTCATTGCTTTC |
| GW8-5DL | WL3957/WL3958 | GACGTACTGTTCTTCAGTAGCC | TCATCTCGTTGTCGTTGGAG |
| IPA1-AS | WL3965/WL3966 | AGGAGGGTTGCACCAGTTG | GCGCAGACTTAGGGTAACGTAG |
| IPA1-7DS | WL3969/WL3970 | TCTTGCTCTCACCGATCACTAC | CCGATACTTCAAATTCATGGTT |
| LP1-3A | WL4051/WL4052 | TAGGAGAATGAGCTGTTTTGAG | CAATGTCCAGCCGAGGCT |
| LP1-3B | WL3943/WL3944 | GGACGGAGGGAGTATTTGAC | CAAGTGAGAGAACATGGCACT |
| LP1-3D | WL4104/WL3946 | TAGGAGAATGAGCTGTTTCG | ACTGGTTTTGTTTGATTCCG |
| TGW6-3.1-3AL | WL4053/WL3984 | CTCCATGAGCCGGTTGGT | TGGACGGACGAATCCACTACTA |
| TGW6-3.1-3BL | WL3985/WL3986 | CGGGCCCAAACACTTACC | CGATCCAGTGCCTTATCAAC |
| TGW6-3.1-3DL | WL3987/WL3988 | AAGGAAATCCGTCGGGGA | TCTTTCCATCACCTCAGCG |
| TGW6-3.3-3AL | WL3991/WL3992 | AAACCCACTCGGCGTCAT | AATGCCTTAGTAGCTTACATGGG |
| TGW6-3.3-3BL | WL4099/WL3994 | GGATCAGTGCTACTTGCTAT | CTCCTCGACTATGTTCCCATCA |
| TGW6-3.3-3DL | WL3995/WL3996 | TAACCAAGAGTCCAAGAGCCAAG | CAATAGTCCATCCGACGAAAAG |
| TGW6-7.1-7AS | WL4015/WL4016 | TGAATCCATCAAACCAAACCATA | TTCAATTCTGGGAAGGGAAGAAC |
| TGW6-7.1-7DS | WL4017/WL4018 | CGCGCCGGGTATAAACTT | CCTGTTGGTGAAGCGCAA |
| TGW6-7.4-7AS | WL4145/WL4146 | AGTGTCCATGCAAGTTTGAA | TGACCAGCCGAGCTTGTG |
| TGW6-7.4-7BS | WL4110/WL4111 | GACCAGGTCGAATCGGAACTAC | GATGACGAGATGTGTCCGA |
| TGW6-7.4-7DS | WL4027/WL4028 | AAAGAAACCATGAAAAGGAATCG | TTTCAAGCTCACACATCTGCT |
You can download all the above sequences as a single file from here.
We have developed an Agrobacterium-delivered CRISPR/Cas9 system for wheat genome editing. We tested this system in wheat protoplasts using a GFP reporter assay, which includes a 1-bp insertion in GFP coding region that disrupted the protein function, wheat codon-optimized Cas9, and a guide RNA targeting the 1-bp insertion. The protoplast assay not only demonstrated functionality of the Cas9 but also selected U6.1 and U6.3 promoter for driving guide RNA cassettes. Using this CRISPR/Cas9 system, we developed 13 constructs for editing 11 grain-regulatory genes. So far we have identified 64 mutation in T0, T1 and T2 generations for TaCKX2-1, TaGLW7, TaGW2 and TaGW8. Analysis of the mutations showed that majority of them are due to deletions.
We also developed a PCR-Cas9 RNP assay protocol for rapid and accurate detection of edit mutations.