Sea Wheatgrass Genome Project
Dissecting the sea wheatgrass genome to transfer biotic stress resistance and abiotic stress tolerance into wheat.
- PD: Li, Wanlong (South Dakota State University, Brookings, SD)
- Co-PD: Xu, Steven S. (USDA-ARS, Fargo, ND)
- Co-PD: Langham, Marie A. C. (South Dakota State University, Brookings, SD)
- Co-PD: Ma, Qin (South Dakota State University, Brookings, SD)
Wheat production is facing numerous challenges from biotic and abiotic stresses. Alien gene transfer has been an effective approach for wheat germplasm enhancement. Sea wheatgrass (SWG) is a distant relative of wheat and a relatively untapped source for wheat improvement. We have identified high tolerance to waterlogging, manganese toxicity, salinity, heat and low nitrogen and resistance to wheat streak mosaic virus (temperature-insensitive), Fusarium head blight and sawflies (due to solid stem) in SWG and developed a large number of SWG-derived populations.
To facilitate simultaneous discovery and transfer of quantitative trait loci (QTL) for biotic stress resistance and abiotic stress tolerance more efficiently, we propose to dissect the SWG genome with three objectives:
- Develop a draft SWG genome assembly and genome-specific markers,
- Dissect SWG chromosomes into RobTs and localize agriculturally important genes and QTL, and
- Dissect SWG chromosome arms by homoeologous recombination and select sub-arm introgressions of biotic stress resistance and abiotic stress tolerance.
This project addresses the Program Area Priorities of A1141 and NIFA-Kansas Wheat Commission co-fund and will serve our long-term goal to broaden the wheat genetic basis and develop novel germplasm that will contribute to a more sustainable wheat industry.
We are a team of interdisciplinary expertise in cytogenetics, molecular genetics, genomics, bioinformatics, plant breeding and pathology, and have materials, technologies and resources in place as evidenced from strong preliminary results. We expect to deliver a complete package of novel germplasm and associated resources, tools and knowledge for breeding wheat tolerance to abiotic and biotic stress.
Sea Wheat Grass chromosome specific PCR markers generated from this study
| Marker Name | Forward primer (5`-> 3`) | Reverse primer (5`-> 3`) | Homeologous Group | |
|---|---|---|---|---|
| WL4503 | TACAACAGCAACAGAAGTGT | AGACACAGCTCCTCATCAACG | 7 | |
| WL4505 | GTAAGCTGCTCTCACCTGCAT | GGCCCATAAACTGGAACCCT | 7 | |
| WL4511 | TGCCTACACAAAGATGGAAGC | GCTCTGCAATTCTGCTTGTT | 3 | |
| WL4513 | GCTGTTTGCATTCACTTGCT | TTGTGTGAGTTTATGTGTGTGTG | 3 | |
| WL4515 | AAGTTCTAGGGGCAAAAAGGA | CTACGGAGATGCCGATCAA | 4 | |
| WL4517 | ATCCAACTAATTTTGTAAGCGTTAGC | CTACGAGAGCAAGCAGGAGG | 5 | |
| WL4523 | GCAGAACCATCGCCATCTC | GGGAAGCGGACTCTAAAAGAA | 4 | |
| WL4531 | ATATGGAAGAATAACGAACAGCA | GGTTCGGATTGCAGGGTT | 6 | |
| WL4533 | ATATGGAAGAATAACGAACAGCA | GGGCCTTTATGAATTGGCT | 6 | |
| WL4537 | AGATTTGCACTAACACGGGAA | TCGTTCCCCTCGCTTGTAGT | 5 | |
| WL4539 | ATCCAGGGGGTAAGCAACA | GTGCTGAATCGGTGTGGTTT | 4 | |
| WL4569 | GAGGCAGCACATCTAAAACAGC | GCAGAGAGAAAACCAGCTTCA | 1 | |
| WL4581 | GCAGGGAGAAATCAAAGAGGAA | TCTAATAAACCGCACGCAAATA | 1 | |
| WL4583 | CAAAGAAAAAGCTATCACTGGGT | ATTTGCGTGGCTGGTGAA | 1 | |
| WL4591 | CTTGCCCCGTCGATTTAC | ATTGTTAATGGTGGATGAGAAAG | 2 | |
| WL4593 | TTAATGCTGGAAGACGCC | TTTCCTTTGCGGATAGCAGATAG | 2 | |
| WL4619 | GTTATGGGTGTCAGTGAGGTG | GTGAACATCGCTGGAATTGG | 2 | |
| WL4623 | GCGAATAACGGACATCACTAGC | GATCGAGGTCATGGTACACTTGG | 7 | |
| WL4625 | TATGGCTTCATTACCTGTGTTG | CTTTGTTCCCATCATTCTCCTTG | 5 | |
| WL4627 | ACTTGCTGGAAAAACAAGCGATA | GGGACTTTGTTTAGATTTCCAAC | 5 | |
| WL4661 | ATGCTGTGATCGGTTCGGT | TTTTTGGCACGAAACAAACT | 4 | |
| WL4663 | AATGCCTATGAAGACAGGGTAAA | GTTGTGTGGCAGTTGTCGTATAG | 4 | |
| WL4665 | CTTGAAACGGCCCCTACTTTTT | GCTTTTCATGTGCCTAGTTATTG | 2 | |
| WL4830 | TACCATCCAGCCTAACCGAC | CTGGCCCTCTCTATGCACAT | 4 | |
| WL4832 | GGGACGGTACTCCCTCTTCT | TCCAACGGATAGTTCCTGGT | 3 | |
| WL4874 | TAGCCACCAAAAACCTCTCG | GGGGGAGAAACATCAGAAGA | 2 | |
| WL4880 | TAGCACACCTACCACGGACA | GGCAAATAAGGCTGAGGTGA | 2 | |
| WL4902 | AGACCTTGCCGGAATCAAC | CAACCCACATTGCCTTCTCT | 6 | |
| WL4904 | TCTGGAGGCCCACTATTGC | GGTTCGCCAGTTTGCTCTT | 6 | |
| WL4906 | GTCACGCCACCAACCATAG | GGGTCCGATCACATTCACTT | 7 | |
| WL4908 | TTTGCCTCCCAACCATTTAC | ACAACCTCTGCTTCCCTCTG | 1 | |
| WL4914 | TCTTATCCGCACGTTACAGC | GATTTCCCAGGATGCAACAC | 4 | |
| WL4918 | ATCACGACTGAGAGGGGAAA | TTGGTGTGAGCTTGGTTTGA | 6 | |
| WL4922 | TTTTCTTGTTGCTGCGAGTG | GTCCCTTTCGTCACACCCTA | 1 | |
| WL4930 | CAACGGATTTCATCGAGTGT | GTGGTGTTCTTGCTCCCTCT | 5 | |
| WL4934 | TATCCCATTGTCACCACGAA | AGGCTGAGGGTTTGTACCAG | 7 | |
| WL4938 | CATTGCCATTCACAGCATTT | TAGGTTTCGCGGTGAGAATA | 2 | |
| WL4940 | AGAGTTTGGGGCTGAATCCT | TCCACCAAGGTCAGAACACA | 3 | |
| WL4966 | TTTATGCCCAATCTTGTGTTCAT | CCAAAGCCCTTCCTGCAA | 1 | |
| WL4968 | CGCGAATCGTCTTGCTGAAA | AGTCGAAGTTGCATTCCAGTG | 2 | |
| WL4972 | ACCATGAATCGGGCACAGTA | AGATCAGATTACACACCTGCAA | 4 | |
| WL4974 | ACAGCTTGGGTCCATTCTCT | GCTAGTCCACTTAATTCCACCA | 5 | |
| WL4976 | GACTCGTACAAACACCATCTGA | CTGTTCCTCACACCCCTCTA | 6 | |
| WL5009 | GTCGATGTTCAGCTGGCA | GACGAGGGTGGGGAAGTG | 5 | |
| WL5031 | AAGTCCCCATTCCTCAGCAT | TCGCCATCAAAACCCTCTAC | 2 | |
| WL5033 | AATCTCATGTGCGTGCAATG | CACAAAGCTTAACGTGTACTGT | 4 | |
| WL5035 | GGAAAGGCGTCGATGGATAC | GGTAGAGCTGTAGACCGTCG | 4 | |
| WL5039 | CTACTACGAGCGATTGGTGC | CTACTTTATTACCCGGCGCG | 6 | |
| WL5041 | GAATAGGGTTGGCGTCGTTC | TCTGGGTCTGAACTTGCACT | 7 | |
| WL5043 | GAGGGTGTGGTAGGCTTAGTAA | GCATAACAAAGCATCGTCTGT | 7 | |
| WL5049 | CCCACCCGTATCCATCCTC | GCCGAAGAGCAGGAAGGT | 1 | |
| WL5057 | AGTCCTCTTCCTCATGCCAG | GTCCGCCTAGCCTTCAGTT | 3 | |
| WL5059 | GGTAAGTTGGTAACCCTGTAAGT | CTCGTTCAAGGCCAGTATTTGA | 4 | |
| WL5063 | AGAAAAGCCACACACACACA | CGACCCAGAACGACACCAT | 1 | |
| WL5065 | GGACAGCAGGACACAAAGTAAG | TGTTCTTCATTTTACCCCAGTGA | 1 | |
| WL5067 | CCCTACACGCCTGTTTTGTC | TGCAGAGTCACAGCCTTACA | 6 | |
| WL5071 | CGTTTATGTGCTGGATCGGG | GGCCCAGCTTGTCATTTTGA | 7 | |
| WL5073 | GCTCATTCGCAAATTCACCG | GCAGACCGGCCAAATCAA | 7 | |
| WL5079 | TGTAAACACCTAACAACCGCT | AATGCGCCATACAACTCAGG | 3 | |
| WL5081 | TGAACTTGAAGTGGATGCTCA | AGGAACGAGACAACATCACATAT | 3 | |
| WL5083 | GACGGAGAGCGGAGTGTACC | TTTTGCTCCATACCATCCACCT | 5 | |
| WL5085 | TGGAATTACAAATGGATCGATGG | TCATTCAGAAAGCTTGGACAATA | 5 | |
| WL5087 | TGAACTGAAGAGTGCAGAGCAAG | CGAGCGTTCTTTGTATTTGATTT | 1 | |
| WL5091 | CATCTCATTATCACCACTGTTCG | GGGCCTGATGCTAATCTCTT | 2 | |
| WL5093 | GTCGGTTCCTTGATTGCGTC | AGTAAGATTTGACTGCGTGCA | 3 | |
| WL5123 | GTCCGCCACAACGTATGA | TCGAGACAACATTCAGCGAA | 4 | |
| WL5125 | AGCAGAAAGAGAGAGGTGAAGT | GAGCATCCTAGTCAAAGTAACCA | 4 | |
| WL5127 | ACAACATGAGATGCACAGCT | GACTGCGTTGGTGGCTATAC | 3 | |
| WL5129 | TCGAGCGAGCCTTACGATTC | GGGGCTGCAAACGTTCTTAA | 3 | |
| WL5133 | CGATACATAGCACCCTCCGT | CTTTGCTTAGCCAGGTCCAC | 4 | |
| WL5135 | ACCCGTAACGAACACTTGATG | CTCACCCGTTTCTGCGTC | 3 | |
| WL5137 | TTGTGGAATGATCAGTAAACGC | ACACACACCCCAATTCCTGT | 5 | |
| WL5166 | GCGATTGGTGCAAGTAAACT | TCATGTTCAGCCCAGATTAGC | 1 | |
| WL5170 | CCCTTCTTCGACCTACAGCT | GTGATCGACGGGGCTAGAG | 5 | |
| WL5172 | ATAACGCTAGAGACTCCGCC | GAGATCTACGCCACGACTGA | 5 | |
| WL5174 | GTGCAAGCGTCGGTGAATT | TGGTTTATTCGTTTTCAGCCGT | 7 | |
| WL5312 | CTGCTTGGTCGCTGTATGTC | CTCGATCAACCGCTTACCAC | 6 | |
| WL5316 | CAGAGCGACAGATTCAACGG | CTGGAGCCATTGAAGCAGTG | 6 | |
| WL5337 | GATCAGCCCTATCCAAGCAA | AGGTACAAGCAGCCTTAGTGT | 7 | |
| WL5341 | GCCGCTAACCATCCTCAACT | GCCTAGATGACCAGCCCTTA | 7 | |
| WL5356 | TAAGGTCAGGTTGGCTTCGG | TTCGTCGCGGCCCTAAAA | 7 | |
| WL5358 | GCAATCAGGTGTGGGTTTCC | AACTGAGCGAGCTGGATGAT | 7 | |
| WL5384 | TGGTTAGGGTCGTACTCACTG | CGACACGCACGTTATAGAGA | 3 | |
| WL5388 | GGAACACCCTTTCTTCAGTCC | TAGCTATGTGGGCCAAGGAA | 3 | |
| WL5390 | AGAAGATCGACCGCCACAA | GTCGCTGTTGTGCTACTCG | 3 | |
| WL5392 | ACGGGACACTCAATAGAACTGT | GTAAGCCCTAACCCGTTGC | 3 | |
| WL5396 | CCGAATCTGCTGCGAAATAATT | ACCCTTGTCCGTTTAGTTCAC | 2 | |
| WL5400 | CACAGGCAGACTCGCAATT | AGTCACACAGCTCAGTACCC | 2 | |
| WL5406 | TGCGCCCAGTGTATATAGCA | TGCATCTCCCACACTCATCA | 1 | |
| WL5410 | GCGTTTATTGAGCTTCCGGT | TTTTCCTCCGTTCCAGTGTT | 1 | |
| WL5412 | GCCTCTAGAAACTCCCCTCC | GTTTTGAATCGGGCAGCGAT | 1 | |
| WL5414 | ACCGGTTCACCCTTGGAAAA | TGGACTGAGAGGAAGTTCGG | 1 | |
| WL5416 | TCACCTGTTCACCCTTGGAA | TTTGCAGACGATGATTGGGG | 1 | |
| WL5422 | ACCGAATCCAAGCTGATCCT | CTCCTGTTCTGAGTTGGTGC | 1 | |
| WL5428 | CGCCATGTCTACGAAAGCAT | GCAGGCACACATACGATCTC | 7 | |
| WL5430 | TGACAGTGGCGGATCTAGAA | AAGACCCTATCGTGTGGCAG | 1 | |
| WL5432 | GATTTCCGCACTAACTCTTTGC | ACCAGTCATGTACAACCATCAA | 1 | |
| WL5436 | TGGGAATTAATGTGGGGCCT | CGCTCTAAATTTCCCACGGG | 5 | |
| WL5438 | ACCGTCTCACAGCATATGGT | TCGATACCCAGCATCTCCAT | 5 | |
| WL5440 | TAATGTTGGCTTGCATCCGG | ATGACCTCGGCTATGACTGG | 5 |
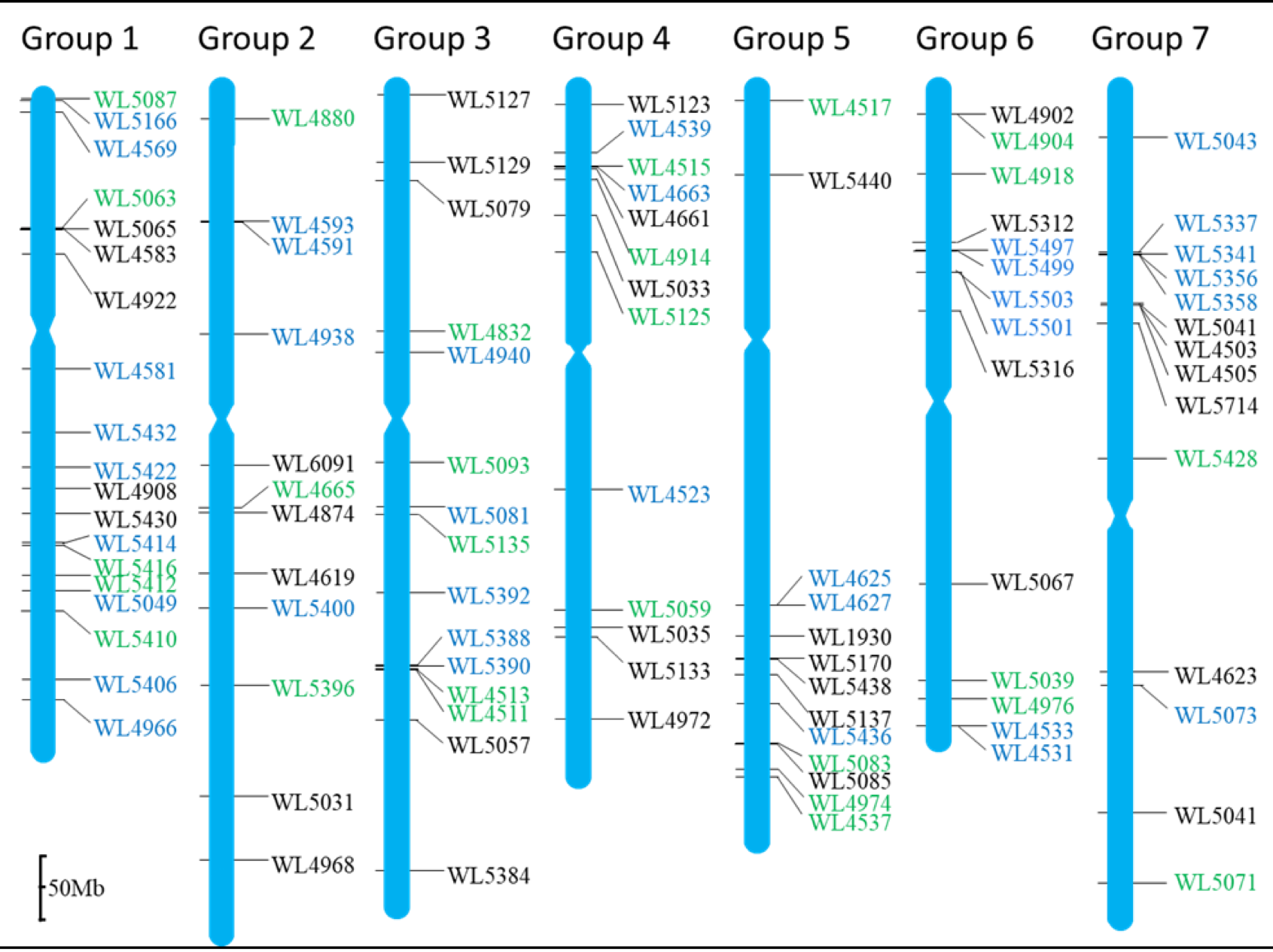
Anchoring the SWG-specific markers to the D-genome chromosomes of Aegilops tauschii and the subgenome origin
The homeologous are indicated at the top of the maps, and the marker loci are indicated at the right of the maps, and their position on the D-genome chromosomes are indicated the black bars. The markers in green are positive for the J1 genome, the markers in the blue are positive for the J2 genome, and the markers in black are codominant for both the J1 and J2 genomes as assayed the wheat-SWG addition lines. The scale bar in the bottom corner indicates 50 Mb.
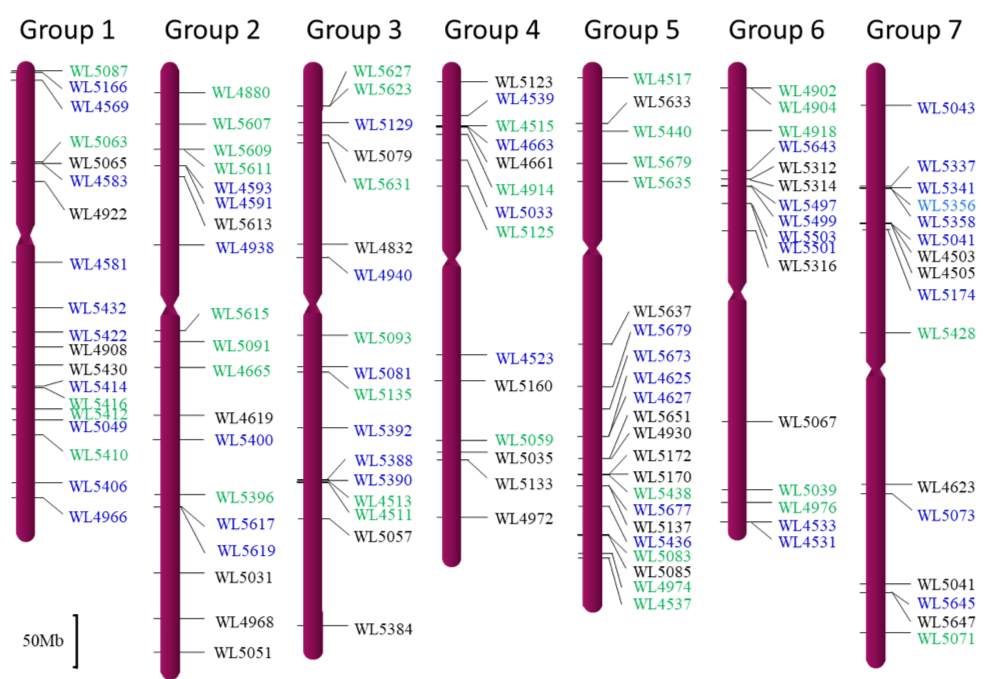
Anchoring the SWG-specific markers to the D-genome chromosomes of Aegilops tauschii
The markers in green are specific to the J1 genome, the markers in blue are specific to the J2 genome, and the markers in black are codominant for both the J1 and the J2 genomes as assayed in the wheat-SWG addition lines. The scale bar in the left bottom corner indicates 50 Mb.
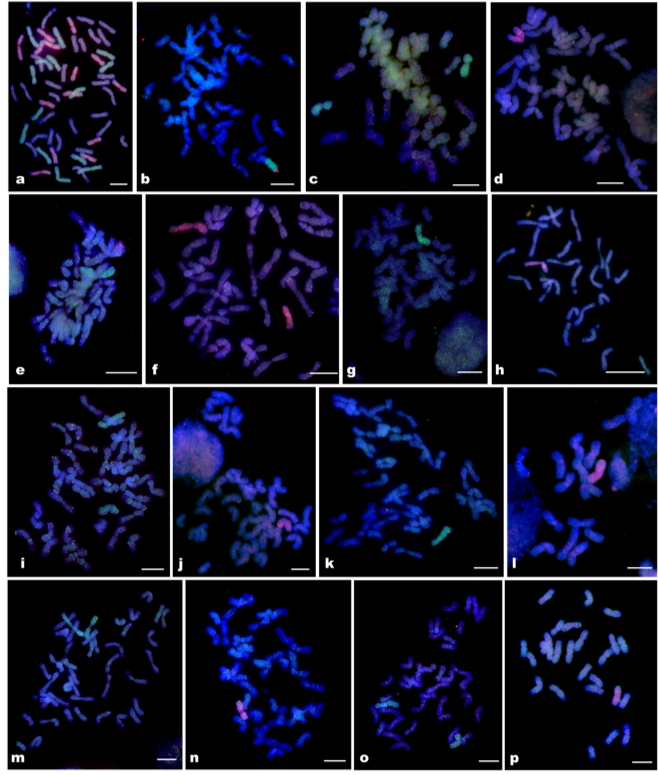
Multi-color GISH analyses of wheat-SWG amphiploid and chromosome lines
a) Amphiploid 13G819, b) 3J1L RobT, c) DA1J1L, d) MA1J2, e) MA2J1, f) MA2J2 + 2J2L, g) MA3J1, h) MA3J2, i) DA4J1, j) MA4J2, k) MA5J1, l) MA5J2, m) DA6J1, n) MA6J2, o) DA7J1, and p) MA7J2. The J1-genome chromosomes were painted in green by biotin-labeled genomic DNA of Th. elongatum, the J2-genome chromosomes were painted in red by digoxigenin-labeled genomic DNA of D. villosum, and the wheat chromosomes were counterstained in blue or purple by DAPI. The scale bars indicate 10 µm.

Assigning traits to SWG chromosomes
a) Segregation of the solid stem (bottom) is associated by chromosome 3J1. b) Segregation of blue seed color (bottom) is associated with chromosome 4J1. c) WSMV resistance (right) is associated with chromosome 2J1. d) WL tolerance (right) is associated with chromosome 1J1.
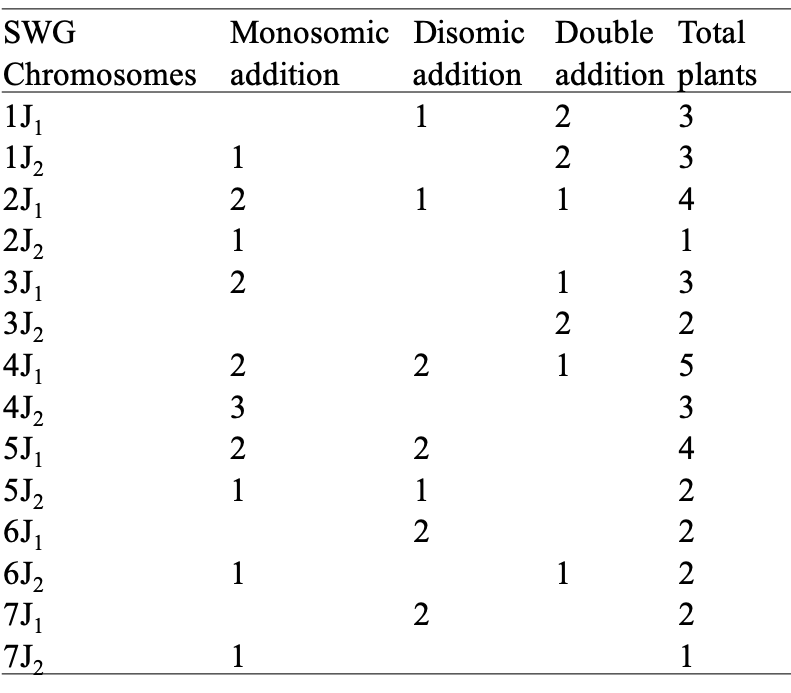
A list of Wheat-SWG addition lines